Multiple Imputation for Missing Data in Meta-Analysis
metami.RdMultiple imputation allows for the uncertainty about the missing data by generating several different plausible imputed data sets and appropriately combining results obtained from each of them. Let \(\hat{\theta}_{*m}\) be the estimated coefficient from the \(m\)th imputed dataset for one of the \(p\) dimensions in the multivariate outcome, where \(m=1,\dots,M\). The coefficient from MI \(\bar{\theta}\) is simply just an arithmetic mean of the individual coefficients estimated from each of the \(M\) meta-analysis. We have $$\bar{\theta}=\frac{\sum_{m=1}^{M}\hat{\theta}_{*m}}{M}.$$ Estimation of the standard error for each variable is little more complicated. Let \(V_W\) be the within imputation variance, which is the average of the variance of the estimated coefficient from each imputed dateset: $$V_W=\frac{\sum_{m=1}^{M}V ({\hat{\theta}_{*m}})}{M},$$ where \(V ({\hat{\theta}_{*m}})\) is the variance of the estimator calculated from generalized least squares methods using the imputed dataset. Let \(V_B\) be the between imputation variance, which is calculated as $$V_B=\frac{\sum_{m=1}^{M}({\hat{\theta}_{*m}}-\bar{\theta})^2}{M-1}.$$ From \(V_W\) and \(V_B\), the variance of the pooled coefficients is calculated as $$V(\bar{\theta})=V_W+V_B+\frac{V_B}{M}$$ The above variance is statistically principled since \(V_W\) reflects the sampling variance and \(V_B\) reflects the extra variance due to the missing data.
metami(data, M = 20, vcov = "r.vcov", r.n.name, ef.name, x.name = NULL, rvcov.method = "average", rvcov.zscore = TRUE, type = NULL, d = NULL, sdt = NULL, sdc = NULL, nt = NULL, nc = NULL, st = NULL, sc = NULL, n_rt = NA, n_rc = NA, r = NULL, func = "mvmeta", formula = NULL, method = "fixed", pool.seq = NULL, return.mi = FALSE, ci.level = 0.95)
Arguments
| data | A \(N \times p\) data frame that contains effect sizes and predictors for meta-regression, if any. |
|---|---|
| M | Number of imputed data sets. |
| vcov | Method for computing effect sizes; options including |
| r.n.name | A string defining the column name for sample sizes in |
| ef.name | A \(p\)-dimensional vector that stores the column names for sample sizes in |
| x.name | A vector that stores the column names in |
| rvcov.method | Method used for |
| rvcov.zscore | Whether the correlation coefficients in |
| type | A \(p\)-dimensional vector indicating types of effect sizes for the argument |
| d | A \(p\)-dimensional vector that stores the column names in |
| sdt | A \(p\)-dimensional vector that stores the column names in |
| sdc | A vector defined in a similar way as |
| nt | A \(p\)-dimensional vector that stores the column names in |
| nc | A vector defined in a similar way as |
| st | A \(p\)-dimensional vector that stores the column names in |
| sc | A vector defined in a similar way as |
| n_rt | A \(N\)-dimensional list of \(p \times p\) correlation matrices storing sample sizes in the treatment group reporting pairwise outcomes in the off-diagonal elements. See |
| n_rc | A list defined in a similar way as |
| r | A \(N\)-dimensional list of \(p \times p\) correlation matrices for the \(p\) outcomes from the \(N\) studies. See |
| func | A string defining the function to be used for fitting the meta-analysis. Options include |
| formula | Formula used for the function |
| method | Method used for the function |
| pool.seq | A numeric vector indicating if the results are pooled from subsets of the |
| return.mi | Should the |
| ci.level | Significant level for the pooled confidence intervals. The default is 0.05. |
Author
Min Lu
Details
For the imputation phase, this function imports the mice package that imputes incomplete multivariate data by chained equations. The pooling phase is performed via the Rubin's rules.
Value
A data.frame that contains the pooled results from the M imputed data sets.
A \(M\)-dimensional list of results from each imputed data set.
A \(M\)-dimensional list of imputed data sets if the argument return.mi = TRUE.
A list of results from the pooled results from the subsets of the M imputed data sets if the argument pool.seq = TRUE.
References
Ahn, S., Lu, M., Lefevor, G.T., Fedewa, A. & Celimli, S. (2016). Application of meta-analysis in sport and exercise science. In N. Ntoumanis, & N. Myers (Eds.), An Introduction to Intermediate and Advanced Statistical Analyses for Sport and Exercise Scientists (pp.233-253). Hoboken, NJ: John Wiley and Sons, Ltd.
Cooper, H., Hedges, L.V., & Valentine, J.C. (Eds.) (2009). The handbook of research synthesis and meta-analysis. New York: Russell Sage Foundation.
Olkin, I., & Ishii, G. (1976). Asymptotic distribution of functions of a correlation matrix. In S. Ikeda (Ed.), Essays in probability and statistics: A volume in honor of Professor Junjiro Ogawa (pp.5-51). Tokyo, Japan: Shinko Tsusho.
Van Buuren, S. and Groothuis-Oudshoorn, K., 2011. mice: Multivariate imputation by chained equations in R. Journal of statistical software, 45(1), pp.1-67.
Gasparrini A., Armstrong, B., Kenward M. G. (2012). Multivariate meta-analysis for non-linear and other multi-parameter associations. Statistics in Medicine. 31(29):3821-3839.
Cheung, M.W.L. (2015). metaSEM: An R Package for Meta-Analysis using Structural Equation Modeling. Frontiers in Psychology 5, 1521.
Rubin, D.B., 2004. Multiple imputation for nonresponse in surveys (Vol. 81). John Wiley & Sons.
Examples
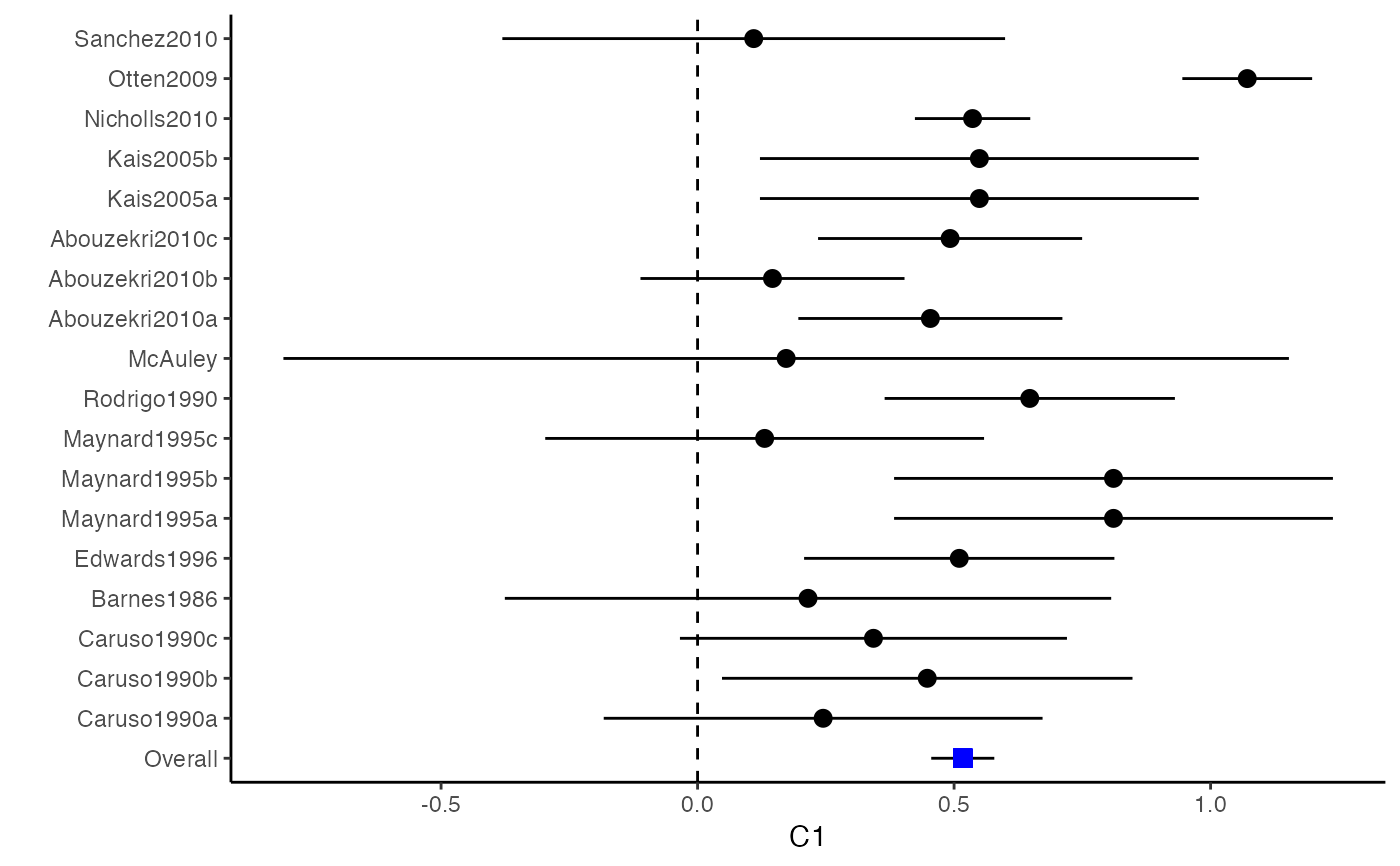
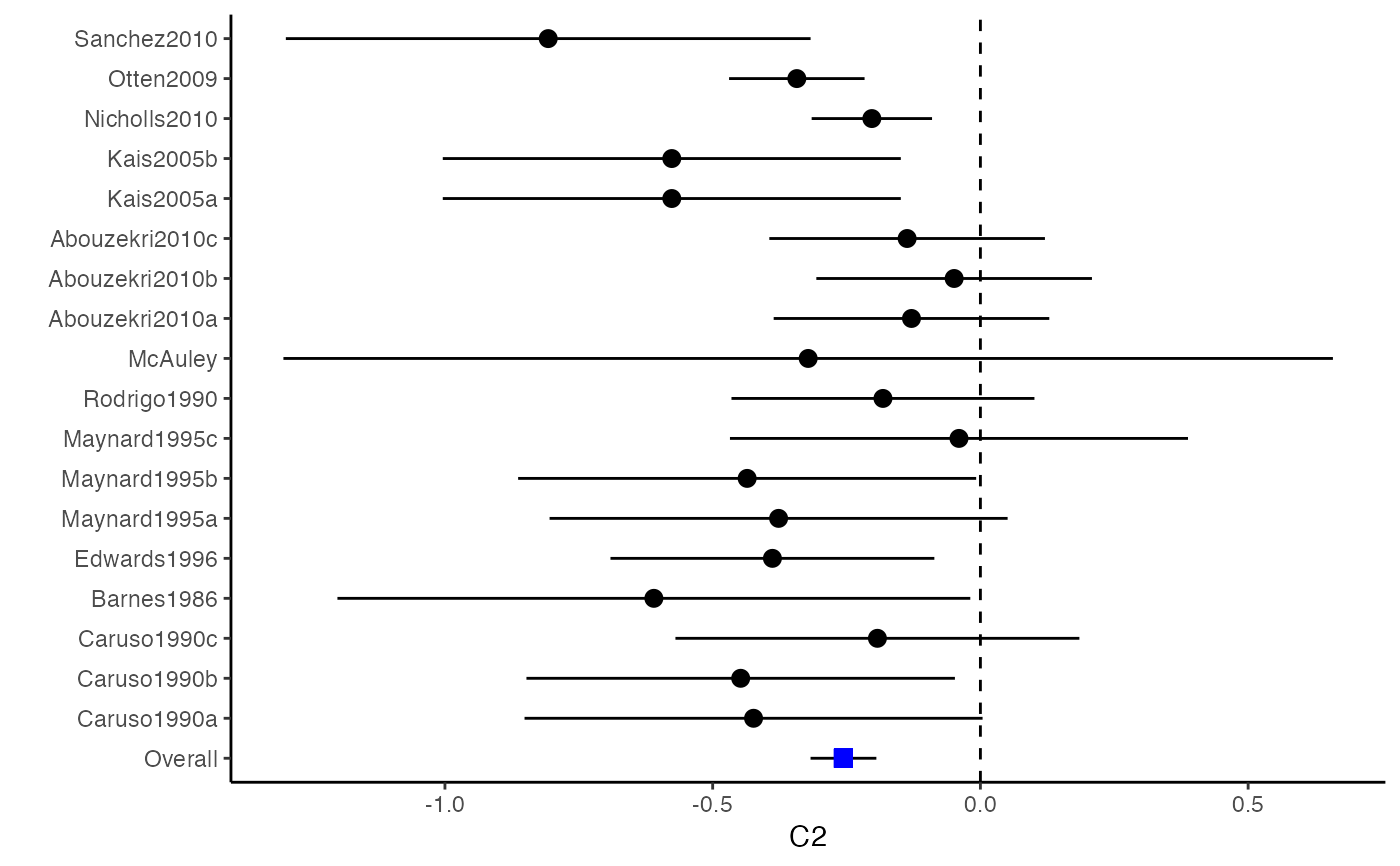
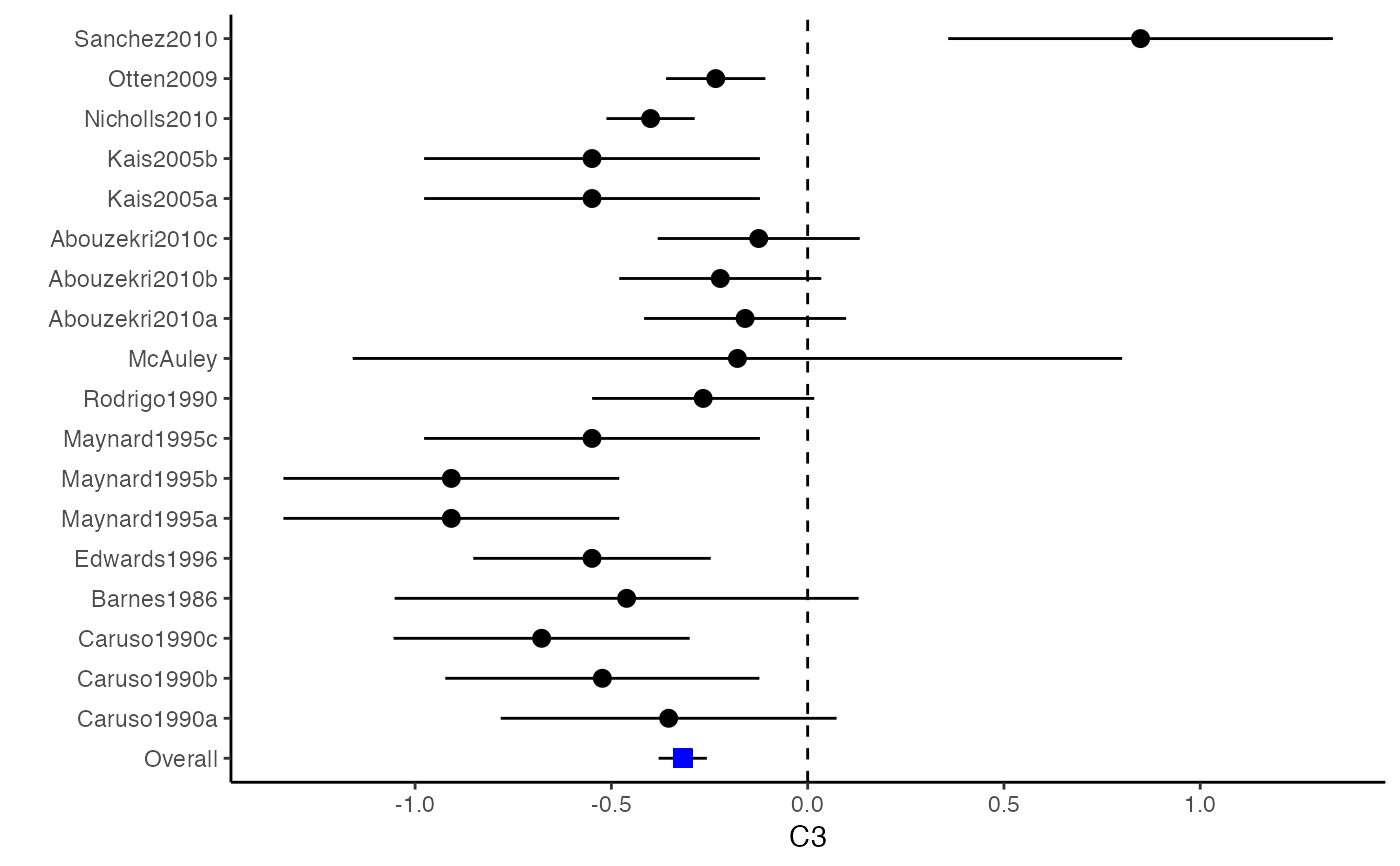
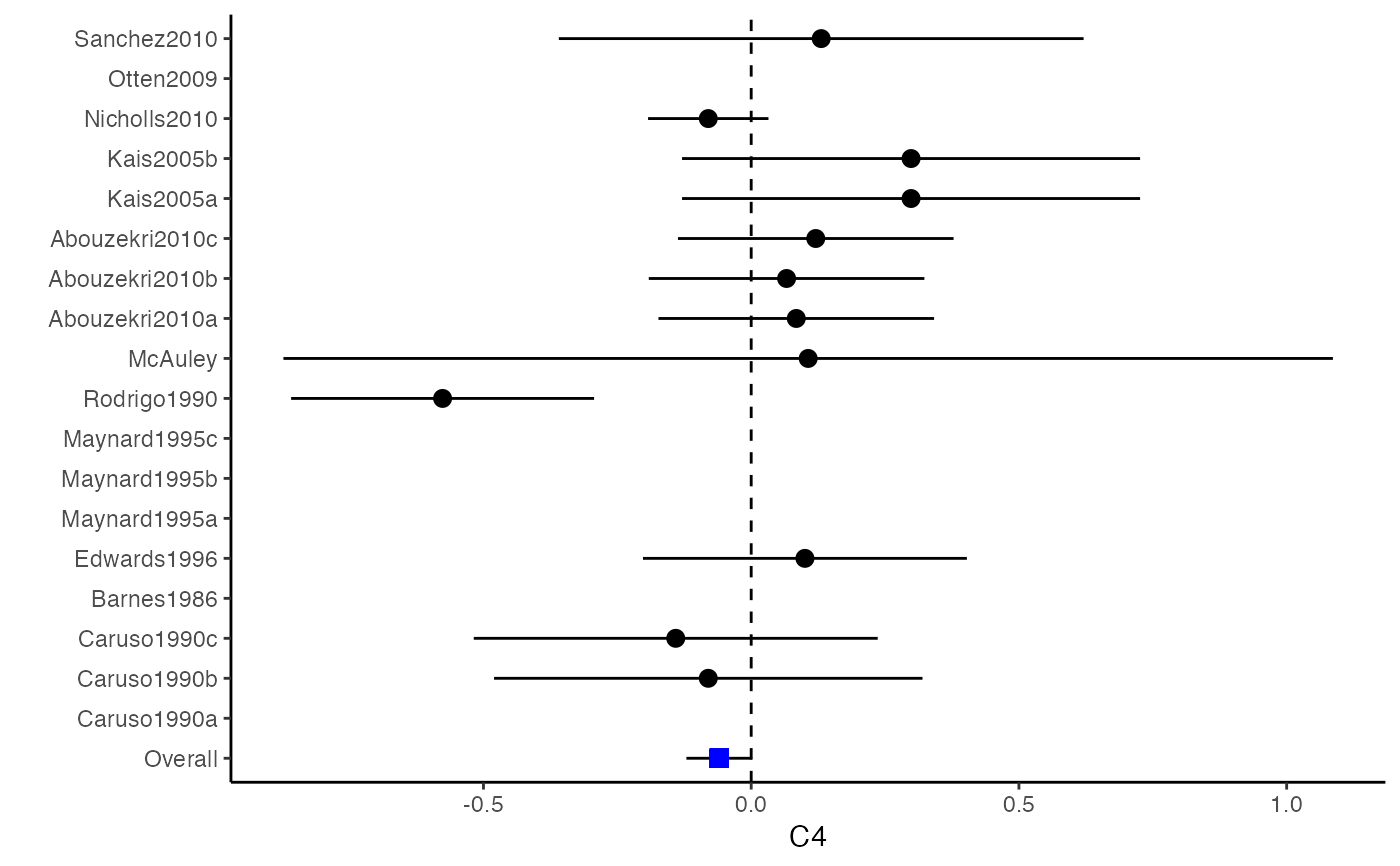
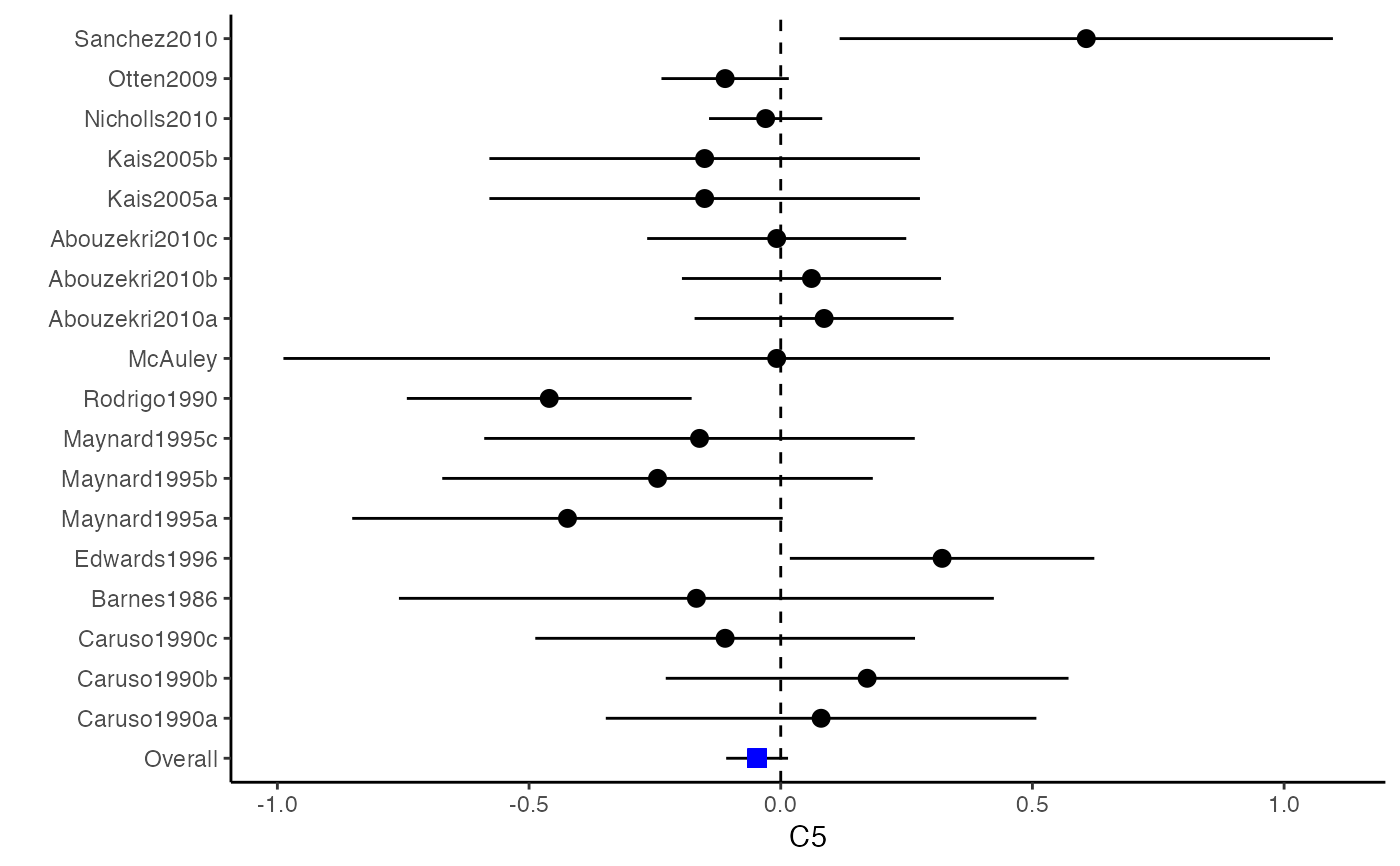
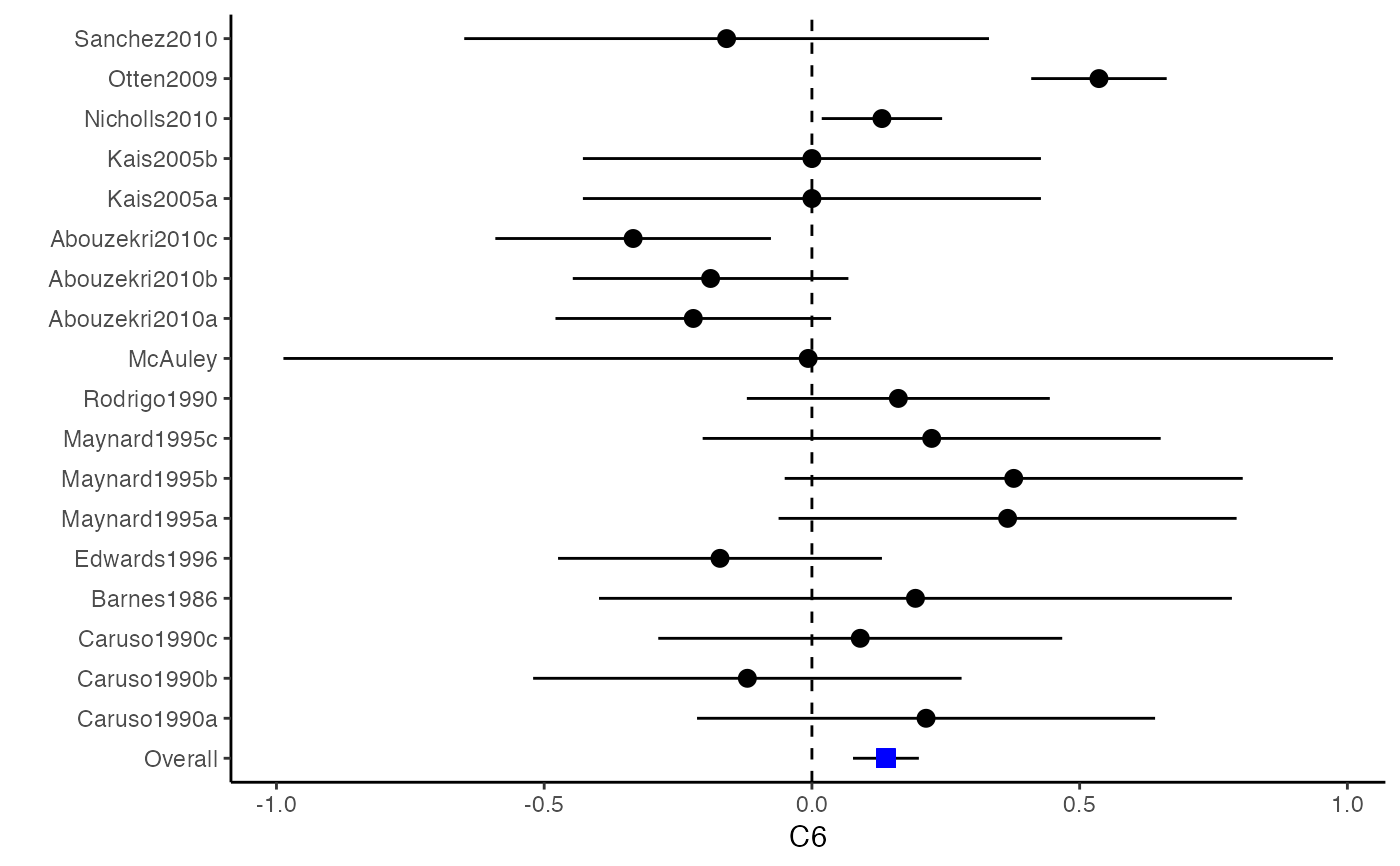
##################################################################################### # Example: Craft2003 data # Preparing input arguments for meta.mi() and fixed-effect model ##################################################################################### # prepare a dataset with missing values and input arguments for meta.mi Craft2003.mnar <- Craft2003[, c(2, 4:10)] Craft2003.mnar[sample(which(Craft2003$C4 < 0), 6), "C4"] <- NA dat <- Craft2003.mnar n.name <- "N" ef.name <- c("C1", "C2", "C3", "C4", "C5", "C6") # fixed-effect model obj <- metami(dat, M = 10, vcov = "r.vcov", n.name, ef.name, func = "metafixed")#> pooled results from 10 imputations for missing values in C4 #> Fixed-effects coefficients #> Estimate Std. Error z Pr(>|z|) 95%ci.lb 95%ci.ub #> C1 0.5169 0.0314 16.4814 0.0000 0.4555 0.5784 *** #> C2 -0.2558 0.0314 -8.1561 0.0000 -0.3172 -0.1943 *** #> C3 -0.3182 0.0314 -10.1467 0.0000 -0.3796 -0.2567 *** #> C4 -0.0596 0.0314 -1.9004 0.0574 -0.1211 0.0019 . #> C5 -0.0469 0.0314 -1.4971 0.1344 -0.1084 0.0145 #> C6 0.1382 0.0314 4.4072 0.0000 0.0768 0.1997 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #>######################## # Plotting the result ######################## computvcov <- r.vcov(n = Craft2003$N, corflat = subset(Craft2003.mnar, select = C1:C6), method = "average") plotCI(y = computvcov$ef, v = computvcov$list.vcov, name.y = NULL, name.study = Craft2003$ID, y.all = obj$coefficients[,1], y.all.se = obj$coefficients[,2])#> $`Plotting C1`#> #> $`Plotting C2`#> #> $`Plotting C3`#> #> $`Plotting C4`#> Warning: Removed 6 rows containing missing values (geom_pointrange).#> #> $`Plotting C5`#> #> $`Plotting C6`#>######################## # Pooling from subsets ######################## o1 <- metami(dat, M = 10, vcov = "r.vcov", n.name, ef.name, func = "metafixed", pool.seq = c(5, 10))#> pooled results from 10 imputations for missing values in C4 #> Fixed-effects coefficients #> Estimate Std. Error z Pr(>|z|) 95%ci.lb 95%ci.ub #> C1 0.5170 0.0314 16.4825 0.0000 0.4555 0.5784 *** #> C2 -0.2560 0.0314 -8.1642 0.0000 -0.3175 -0.1945 *** #> C3 -0.3182 0.0314 -10.1458 0.0000 -0.3796 -0.2567 *** #> C4 -0.0501 0.0314 -1.5976 0.1101 -0.1116 0.0114 #> C5 -0.0470 0.0314 -1.4998 0.1337 -0.1085 0.0144 #> C6 0.1382 0.0314 4.4058 0.0000 0.0767 0.1997 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #># pooled results from M = 5 imputed data sets o1$result.seq$M5$coefficients#> Estimate Std. Error z Pr(>|z|) 95%ci.lb 95%ci.ub #> C1 0.51683598 0.03136417 16.4785456 0.000000e+00 0.45536333 0.57830863 #> C2 -0.25631789 0.03135624 -8.1743812 2.220446e-16 -0.31777500 -0.19486078 #> C3 -0.31804815 0.03135804 -10.1424747 0.000000e+00 -0.37950878 -0.25658751 #> C4 -0.01607465 0.03135730 -0.5126286 6.082112e-01 -0.07753384 0.04538454 #> C5 -0.04727197 0.03135605 -1.5075871 1.316602e-01 -0.10872869 0.01418475 #> C6 0.13815009 0.03136602 4.4044512 1.060520e-05 0.07667382 0.19962635# pooled results from M = 10 imputed data sets o1$result.seq$M10$coefficients#> Estimate Std. Error z Pr(>|z|) 95%ci.lb 95%ci.ub #> C1 0.51696741 0.03136463 16.482497 0.000000e+00 0.45549387 0.57844096 #> C2 -0.25600261 0.03135676 -8.164193 2.220446e-16 -0.31746073 -0.19454450 #> C3 -0.31815427 0.03135835 -10.145761 0.000000e+00 -0.37961549 -0.25669304 #> C4 -0.05009895 0.03135803 -1.597643 1.101224e-01 -0.11155955 0.01136166 #> C5 -0.04702938 0.03135615 -1.499845 1.336544e-01 -0.10848629 0.01442754 #> C6 0.13819176 0.03136610 4.405768 1.054095e-05 0.07671534 0.19966818######################################################################################### # Running random-effects and meta-regression model using packages "mvmeta" or "metaSEM" ######################################################################################### # Restricted maximum likelihood (REML) estimator from the mvmeta package # library(mvmeta) # o2 <- metami(dat, M = 10, vcov = "r.vcov", # n.name, ef.name, # formula = as.formula(cbind(C1, C2, C3, C4, C5, C6) ~ . ), # func = "mvmeta", # method = "reml") # maximum likelihood estimators from the metaSEM package # library(metaSEM) # o3 <- metami(dat, M = 10, vcov = "r.vcov", # n.name, ef.name, # func = "meta") # meta-regression # library(metaSEM) # o4 <- metami(dat, M = 10, vcov = "r.vcov", # n.name, ef.name, x.name = "p_male", # func = "meta") # library(mvmeta) # o5 <- metami(dat, M = 20, vcov = "r.vcov", # n.name, ef.name, x.name = "p_male", # formula = as.formula(cbind(C1, C2, C3, C4, C5, C6) ~ p_male ), # func = "mvmeta", # method = "reml") ##################################################################################### # Example: Geeganage2010 data # Preparing input arguments for meta.mi() and fixed-effect model ##################################################################################### # Geeganage2010.mnar <- Geeganage2010 # Geeganage2010.mnar$MD_SBP[sample(1:nrow(Geeganage2010),7)] <- NA # r12 <- 0.71 # r13 <- 0.5 # r14 <- 0.25 # r23 <- 0.6 # r24 <- 0.16 # r34 <- 0.16 # r <- vecTosm(c(r12, r13, r14, r23, r24, r34)) # diag(r) <- 1 # mix.r <- lapply(1:nrow(Geeganage2010), function(i){r}) # o <- metami(data = Geeganage2010.mnar, M = 10, vcov = "mix.vcov", # ef.name = c("MD_SBP", "MD_DBP", "RD_DD", "lgOR_D"), # type = c("MD", "MD", "RD", "lgOR"), # d = c("MD_SBP", "MD_DBP", NA, NA), # sdt = c("sdt_SBP", "sdt_DBP", NA, NA), # sdc = c("sdc_SBP", "sdc_DBP", NA, NA), # nt = c("nt_SBP", "nt_DBP", "nt_DD", "nt_D"), # nc = c("nc_SBP", "nc_DBP", "nc_DD", "nc_D"), # st = c(NA, NA, "st_DD", "st_D"), # sc = c(NA, NA, "sc_DD", "sc_D"), # r = mix.r, # func = "metafixed")