Computing Variance-Covariance Matrices for Standardized Mean Differences
smd.vcov.RdThe function lgOR.vcov computes effect sizes and variance-covariance matrix for multivariate meta-analysis when the effect sizes of interest are all measured by standardized mean difference. See mix.vcov for effect sizes of the same or different types.
smd.vcov(nt, nc, d, r, n_rt = NA, n_rc = NA, name = NULL)
Arguments
| nt | A \(N \times p\) matrix storing sample sizes in the treatment group reporting the \(p\) outcomes. |
|---|---|
| nc | A matrix defined in a similar way as |
| d | A \(N \times p\) matrix or data frame with standard mean differences (SMD) from the \(N\) studies. |
| r | A \(N\)-dimensional list of \(p \times p\) correlation matrices for the \(p\) outcomes from the \(N\) studies. |
| n_rt | A \(N\)-dimensional list of \(p \times p\) matrices storing sample sizes in the treatment group reporting pairwise outcomes in the off-diagonal elements. |
| n_rc | A list defined in a similar way as |
| name | Names for the outcomes. |
Author
Min Lu
Value
| ef | A \(N \times p\) data frame that transforms the input argument d into Hedges's g (Wei and Higgins, 2013). |
| list.vcov | A \(N\)-dimensional list of \(p(p+1)/2 \times p(p+1)/2\) variance-covariance matrices for Hedges's g (Wei and Higgins, 2013). |
| matrix.vcov | A \(N \times p(p+1)/2\) whose rows are computed variance-covariance vectors for Hedges's g (Wei and Higgins, 2013). |
| list.dvcov | A \(N\)-dimensional list of \(p(p+1)/2 \times p(p+1)/2\) variance-covariance matrices for SMD (Olkin and Gleser, 2009). |
| matrix.dvcov | A \(N \times p(p+1)/2\) matrix whose rows are computed variance-covariance vectors for SMD (Olkin and Gleser, 2009). |
References
Ahn, S., Lu, M., Lefevor, G.T., Fedewa, A. & Celimli, S. (2016). Application of meta-analysis in sport and exercise science. In N. Ntoumanis, & N. Myers (Eds.), An Introduction to Intermediate and Advanced Statistical Analyses for Sport and Exercise Scientists (pp.233-253). Hoboken, NJ: John Wiley and Sons, Ltd.
Wei, Y. & Higgins, J. (2013). Estimating within study covariances in multivariate meta-analysis with multiple outcomes. Statistics in Medicine, 32(7), 119-1205.
Olkin, I. & Gleser, L. (2009). Stochastically dependent effect sizes. In H. Cooper, L. V. Hedges, & J. C. Valentine (Eds.), The handbook of research synthesis and meta-analysis (pp. 357-376). New York: Russel Sage Foundation.
Examples
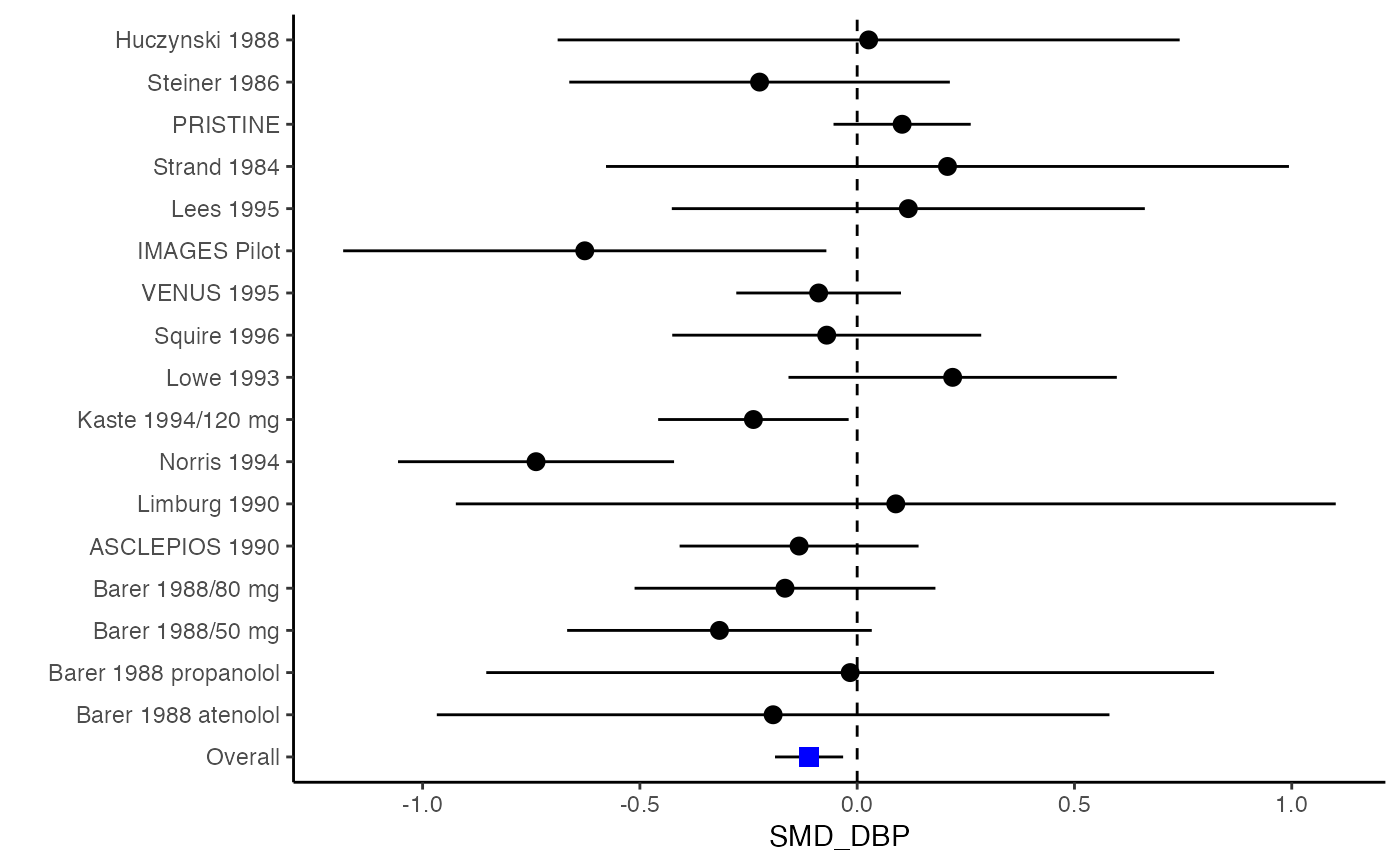
###################################################### # Example: Geeganage2010 data # Preparing covarianceS for multivariate meta-analysis ###################################################### data(Geeganage2010) ## set the correlation coefficients list r r12 <- 0.71 r.Gee <- lapply(1:nrow(Geeganage2010), function(i){matrix(c(1, r12, r12, 1), 2, 2)}) computvcov <- smd.vcov(nt = subset(Geeganage2010, select = c(nt_SBP, nt_DBP)), nc = subset(Geeganage2010, select = c(nc_SBP, nc_DBP)), d = subset(Geeganage2010, select = c(SMD_SBP, SMD_DBP)), r = r.Gee, name = c("SMD_SBP", "SMD_DBP")) # name variance-covariance matrix as S S <- computvcov$matrix.vcov ## fixed-effect model y <- computvcov$ef MMA_FE <- summary(metafixed(y = y, Slist = computvcov$list.vcov)) ####################################################################### # Running random-effects model using package "mvmeta" or "metaSEM" ####################################################################### # Restricted maximum likelihood (REML) estimator from the mvmeta package #library(mvmeta) #mvmeta_RE <- summary(mvmeta(cbind(SMD_SBP, SMD_DBP), # S = S, # data = y, # method = "reml")) #mvmeta_RE # maximum likelihood estimators from the metaSEM package # library(metaSEM) # metaSEM_RE <- summary(meta(y = y, v = S)) # metaSEM_RE ############################################################## # Plotting the result: ############################################################## obj <- MMA_FE # obj <- mvmeta_RE # obj <- metaSEM_RE # pdf("CI.pdf", width = 4, height = 7) plotCI(y = computvcov$ef, v = computvcov$list.vcov, name.y = NULL, name.study = Geeganage2010$studyID, y.all = obj$coefficients[,1], y.all.se = obj$coefficients[,2])#> $`Plotting SMD_SBP`#> #> $`Plotting SMD_DBP`#># dev.off()